Self-learning AI: a boost for digital pathology
Author: Mark Nicholls.
“In comparison, unsupervised self-learning AI does not require labels; the system learns image features without aid; it is rapid; capacity is limited only by content; and is wholly interpretable”
Prof. John Le Quesne
The presentation was given by John Le Quesne, Professor of Molecular Pathology at the University of Glasgow and an Associate Group Leader at the CRUK Research Institute in Glasgow. The expert pointed to the application of self-learning AI in lung adenocarcinoma with H&E (Haematoxylin and Eosin) images and its potential in other modalities, such as data-rich multiplex images.
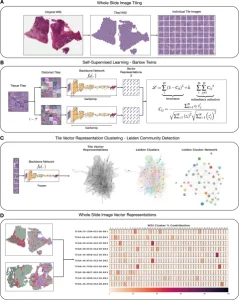
 Focusing on Histomorphological Phenotype Learning (HPL), Le Quesne pointed out the potential of this new approach for discovering meaningful recurrent morphological landscapes in a set of histology images, and assigning ‘interpretable quantitative summary vectors’ to whole slide images (WSI).1 HPL performs particularly well in lung cancer and mesothelioma cohorts, said Le Quesne.
Focusing on Histomorphological Phenotype Learning (HPL), Le Quesne pointed out the potential of this new approach for discovering meaningful recurrent morphological landscapes in a set of histology images, and assigning ‘interpretable quantitative summary vectors’ to whole slide images (WSI).1 HPL performs particularly well in lung cancer and mesothelioma cohorts, said Le Quesne.
Farewell to the “black box”
He contrasted the powerful self-learning AI approach to classical supervised AI where expert pathologists generate labels by annotation of images. ‘The supervised approach is very expensive in terms of time and salary and runs the danger of ending up with a black box which can give strong opinions but might have more difficulty in explaining how it got there,’ he said. ‘In comparison, unsupervised self-learning AI does not require labels; the system learns image features without aid; it is rapid; capacity is limited only by content; and is wholly interpretable.’
Le Quesne illustrated how self-learning AI had been applied in lung adenocarcinoma H&E images that were split into a large number of tiles without any annotation. He said: ‘Tiles then individually go through the deep learning architecture which in an unsupervised way discovers meaningful features of those tile images and can then identify the tiles that are morphologically relevant.’
Photo
From there, H&E images can be converted into a map of histomorphological clusters (HPCs) that deliver interpretable quantitative histopathology and can predict recurrence via that map which contains the classic morphologies for carcinoma and gives a detailed morphological picture. ‘It gives us a really interpretable quantitative tool to classify images and search tissues and we can very quickly view the morphological content of that tissue,’ he continued. ‘Even better, we can use percentage compositions to ask if there are clusters which significantly affect patient outcome. HPC composition is highly prognostic in validation cohorts.’
A potent tool – with some remaining challenges
The expert described it as a ‘potent tool’ that outperforms the pathologist and supervised AI grading. ‘Self-learning methods like HPL learn meaningful pathology that resides in these images without supervision; provides a quantitative interpretable morphological dictionary, automates identification, outperforms supervised methods and human experts in key tasks such as subtyping and prognosis, and is capable of discovering ‘new’ features and morphologies,’ said Le Quesne.
He additionally highlighted how self-learning AI can be deployed in other imaging modalities such as multiplex immunofluorescence (IF) and be applied to the immune microenvironment. ‘With images put through the same process with much smaller tiles, we find ourselves able to prognosticate outcomes really accurately with greater power than human grading of whole slide images,’ he added.
‘Self-learning AI can be applied to multiplex panels which quantify the hallmarks of cancer, and again we find that self-learned morphological clusters predict outcome, while focused panel design lets us link areas of biology to real world outcomes such as response. There is also huge potential in biomarker discovery.’
However, there are still challenges to overcome, such as in patient selection, turnaround time, quality control and refining how the approach fits with existing platforms and workflows.
Profile: John Le Quesne is a Professor of Molecular Pathology at the University of Glasgow, an Associate Group Leader at the CRUK Research Institute in Glasgow, and an honorary consultant pathologist with NHS Greater Glasgow and Clyde. His group uses a combination of classical and novel quantitative pathological methods to generate and test hypotheses about human tumour biology.
Discover our full series of Digital Pathology meetings here.
Mark Nicholls is writer for Health Care in Europe. This Report from the 11th Digital Pathology & AI Congress is reproduced with permission and thanks.






